Protein Synthesis and Genetic Code: Short Notes (Q&A Style)
These notes cover the key components of protein synthesis, the genetic code, its discovery, and the stages of translation (initiation, elongation, termination). Also included are details on tRNA and rRNA structures. Written in simple language like EasyBiologyClass.com, with clear paragraphs and examples.
1. What are the main components involved in protein synthesis?
Protein synthesis, or translation, is the process where cells use genetic instructions to build proteins. The key components include nucleic acids like mRNA (messenger RNA, which carries the code from DNA), tRNA (transfer RNA, which delivers amino acids), and rRNA (ribosomal RNA, which forms the ribosome). Ribosomes are the main site of synthesis, made of rRNA and proteins, with sites like A-site for new tRNA, P-site for the growing chain, and E-site for exit. Amino acids are the building blocks (20 types), activated by enzymes called aminoacyl-tRNA synthetases. Energy comes from ATP and GTP, and factors like initiation, elongation, and release proteins help regulate steps. Magnesium ions stabilize everything. In prokaryotes, it's faster and coupled with transcription; in eukaryotes, it's in the cytoplasm or ER.
2. What is the genetic code, and what are its main features?
The genetic code is the set of rules that translates nucleotide triplets in mRNA (called codons) into specific amino acids during protein synthesis. Each codon is three bases (e.g., AUG for methionine), making 64 possible codons from four bases (A, U, G, C). It is degenerate, meaning multiple codons code for the same amino acid (like four for alanine), which reduces harmful mutation effects. The code is unambiguous (one codon, one amino acid), non-overlapping, and comma-free (read continuously). Start codon AUG begins translation, while stop codons (UAA, UAG, UGA) end it without coding for amino acids. The wobble hypothesis explains how the third base can vary, allowing fewer tRNAs. It is nearly universal across life, with minor exceptions in mitochondria.
3. Who discovered the genetic code, and how was it done?
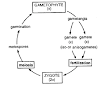
The genetic code was discovered in the 1960s through key experiments. Marshall Nirenberg and J. Heinrich Matthaei started in 1961 using a cell-free E. coli system: they added synthetic poly-U mRNA and radioactive amino acids, finding poly-phenylalanine formed, so UUU codes for phenylalanine. This earned Nirenberg the Nobel Prize in 1968. Har Gobind Khorana built on this by synthesizing repeating copolymers (e.g., poly-UUC) to assign all 64 codons, confirming degeneracy. Francis Crick proposed the triplet nature earlier (1950s) and the wobble hypothesis in 1966. Sydney Brenner helped with frame-shift experiments. By 1967, the full code was mapped, showing its universality from bacteria to humans. This breakthrough enabled modern genetics and biotech.
4. What is the structure of tRNA, and how does it work in protein synthesis?
tRNA is a small adaptor molecule (70-90 nucleotides) that links the genetic code to amino acids. Its secondary structure is a cloverleaf: it has an acceptor stem at the 3' end (with CCA sequence for attaching amino acids via a high-energy bond), an anticodon loop (7 nucleotides with the anticodon triplet that base-pairs with mRNA codons, e.g., UAC for AUG), a D-loop (with dihydrouridine for stability), a TψC loop (for ribosome binding, with pseudouridine), and a variable loop for diversity. In tertiary structure, it folds into an L-shape: one arm holds the amino acid, the other the anticodon, stabilized by magnesium ions and modified bases (about 10-20% modified for accuracy). During translation, charged tRNA (with amino acid) enters the ribosome's A-site, matches the codon, and transfers the amino acid to the chain. One tRNA per amino acid type, but wobble allows one to read multiple codons.
5. What is the structure of rRNA, and what is its role in protein synthesis?
rRNA forms the core of ribosomes, acting as both scaffold and catalyst (a ribozyme). It is highly folded into helices and loops. In prokaryotes (70S ribosome), the small 30S subunit has 16S rRNA (~1,500 nucleotides, 3 domains for mRNA decoding), and the large 50S has 23S rRNA (~2,900 nt, 6 domains with peptidyl transferase center or PTC for peptide bonds) plus 5S rRNA (~120 nt). Eukaryotes (80S) have larger versions: 18S in 40S subunit, and 28S, 5.8S, 5S in 60S. The PTC in the large subunit's domain V catalyzes bonds without proteins. rRNA is processed from larger precursors in the nucleolus (eukaryotes). It binds mRNA/tRNA, moves the ribosome, and ensures accuracy. Cryo-EM studies (Nobel 2009) showed its atomic details, supporting the RNA world idea where RNA evolved first.
6. Describe the initiation stage of protein synthesis.
Initiation assembles the ribosome to start reading mRNA at the AUG codon. In prokaryotes, the small 30S subunit binds mRNA via Shine-Dalgarno sequence near AUG, then initiator tRNA (with formyl-methionine) joins with IF1, IF2-GTP, IF3. The 50S subunit attaches, GTP hydrolyzes, factors release, forming 70S with P-site occupied. In eukaryotes, the 40S subunit with eIFs binds the 5' cap, scans to AUG, Met-tRNA joins with eIF2-GTP, then 60S attaches. It uses 1 GTP and ensures correct start. Errors here can cause diseases like cancer. The process takes seconds, setting the reading frame.
7. What happens during the elongation stage of protein synthesis?
Elongation adds amino acids to grow the polypeptide chain in a cycle. First, aminoacyl-tRNA (with EF-Tu-GTP in prokaryotes or eEF1A-GTP in eukaryotes) binds the A-site codon. GTP hydrolyzes for accuracy. Then, peptidyl transferase (rRNA) forms a peptide bond between P-site and A-site amino acids; the chain transfers to A-site tRNA. Next, translocation: EF-G-GTP (or eEF2) shifts the ribosome 3 nucleotides along mRNA, moving tRNAs from A to P-site and P to E-site (empty tRNA exits). Per amino acid: 2 GTP and ATP from charging. It adds ~20 amino acids/second in bacteria, from N- to C-terminus. Antibiotics like erythromycin block this.
8. Explain the termination stage of protein synthesis.
Termination ends translation when a stop codon (UAA, UAG, UGA) reaches the A-site. No tRNA binds; instead, release factors enter: RF1/RF2 recognize specific stops, RF3-GTP helps. In eukaryotes, eRF1 binds all stops, eRF3-GTP aids. Peptidyl transferase hydrolyzes the bond, releasing the polypeptide from P-site tRNA. The ribosome splits (IF3 or eIFs help), mRNA/tRNAs exit. Uses 1 GTP. The protein folds, gets modified (e.g., phosphorylation), and is targeted (e.g., to ER via signal peptide). Errors cause truncated proteins, linked to diseases like cystic fibrosis.
Related Topics Table (Quick Links)
| Topic | Key Point | Example/Link Idea |
|---|---|---|
| tRNA vs. rRNA | tRNA: Adaptor; rRNA: Catalyst | L-shape vs. helices |
| Prok vs. Euk | 70S free; 80S ER-bound | fMet vs. Met start |
| Genetic Code Table | 64 codons, 20 AA + 3 stops | AUG = Start/Met |
| Discovery Impact | Nirenberg 1961 → Modern biotech | Synthetic genes |
Section 1: Components of Protein Synthesis (MCQs 1-10)
- What is the primary site of protein synthesis in eukaryotic cells? a) Nucleus b) Ribosome c) Mitochondria d) Golgi apparatus Answer: b) RibosomeReason: Ribosomes (80S in eukaryotes) are the cellular factories where mRNA is translated into proteins. They consist of rRNA and proteins, located in cytoplasm or on rough ER.
- Which molecule carries amino acids to the ribosome during translation? a) mRNA b) rRNA c) tRNA d) DNA Answer: c) tRNAReason: tRNA acts as an adaptor, with anticodon matching mRNA codon and amino acid attached at 3' end, delivering building blocks for polypeptide chain.
- What activates amino acids for attachment to tRNA? a) ATP and aminoacyl-tRNA synthetase b) GTP and ribosomes c) NADH and mitochondria d) Poly-A tail Answer: a) ATP and aminoacyl-tRNA synthetaseReason: Aminoacyl-tRNA synthetases (one per amino acid) use ATP to form ester bond between amino acid and tRNA's CCA end, charging it for translation.
- In which site of the ribosome does the initiator tRNA bind during initiation? a) A-site b) P-site c) E-site d) T-site Answer: b) P-siteReason: P-site (peptidyl) holds the growing chain or initiator tRNA (Met or fMet); A-site is for incoming tRNA, E-site for exit.
- What provides energy for tRNA charging in protein synthesis? a) ATP only b) GTP only c) ATP and GTP d) NADH Answer: c) ATP and GTPReason: ATP for amino acid activation; GTP for initiation factors, elongation (EF-Tu, EF-G), and termination.
- Which ion stabilizes the ribosome structure? a) Na⁺ b) K⁺ c) Mg²⁺ d) Ca²⁺ Answer: c) Mg²⁺Reason: Mg²⁺ ions bridge negatively charged phosphate backbones in rRNA, maintaining ribosomal folding and function.
- What is the role of rRNA in ribosomes? a) Carries genetic code b) Forms scaffold and catalyzes peptide bonds c) Delivers amino acids d) Transcribes DNA Answer: b) Forms scaffold and catalyzes peptide bondsReason: rRNA is a ribozyme; its peptidyl transferase center (PTC) in large subunit catalyzes amide bond formation without proteins.
- In prokaryotes, what sequence on mRNA helps ribosome binding? a) 5' cap b) Shine-Dalgarno sequence c) Poly-A tail d) Kozak sequence Answer: b) Shine-Dalgarno sequenceReason: Shine-Dalgarno (upstream of AUG) base-pairs with 16S rRNA in 30S subunit, positioning for initiation in bacteria.
- Which component ensures fidelity in codon-anticodon pairing? a) Wobble base b) Aminoacyl-tRNA synthetase c) Release factors d) Elongation factors Answer: b) Aminoacyl-tRNA synthetaseReason: These enzymes proofread and attach correct amino acid to tRNA, preventing errors (1 in 10,000 accuracy).
- What is the eukaryotic equivalent of prokaryotic fMet-tRNA? a) Met-tRNA with 5' cap b) Unmodified Met-tRNA c) Formyl-Met-tRNA d) Ala-tRNA Answer: b) Unmodified Met-tRNAReason: Eukaryotes use plain Met-tRNAi (initiatior) for AUG; often cleaved post-translation, unlike formylated in prokaryotes.
Section 2: Genetic Code (MCQs 11-20)
- How many codons are there in the genetic code? a) 20 b) 61 c) 64 d) 3 Answer: c) 64Reason: 4 bases (A,U,G,C) in triplets: 4³ = 64 codons (61 for amino acids + 3 stops).
- What property of the genetic code allows multiple codons for one amino acid? a) Universality b) Degeneracy c) Ambiguity d) Overlapping Answer: b) DegeneracyReason: Redundancy (e.g., 6 for Leu) buffers mutations; usually third base varies.
- Which codon initiates translation? a) UGA b) AUG c) UAA d) UAG Answer: b) AUGReason: AUG codes for Met (or fMet) and signals start; sets reading frame.
- Which of the following is a stop codon? a) GGG b) UGA c) AUU d) CCC Answer: b) UGAReason: UGA (amber), UAA (ochre), UAG (opal) have no tRNA; bind release factors to end translation.
- What does the wobble hypothesis explain? a) Start codons b) Third base flexibility in anticodon c) Overlapping genes d) Mutations Answer: b) Third base flexibility in anticodonReason: Proposed by Crick (1966); e.g., U in anticodon pairs with A or G, reducing tRNA need to ~32.
- The genetic code is read in which direction? a) 5' to 3' b) 3' to 5' c) Bidirectional d) Random Answer: a) 5' to 3'Reason: mRNA is synthesized/transcribed 5'→3'; translation follows same direction, non-overlapping.
- Which amino acid is coded by only one codon? a) Leucine b) Tryptophan c) Serine d) Arginine Answer: b) TryptophanReason: UGG is unique for Trp; Met (AUG) is start but shared; others have 2-6.
- The genetic code is nearly universal except in: a) Bacteria b) Mitochondria c) Plants d) Fungi Answer: b) MitochondriaReason: Mitochondrial code varies (e.g., AUA=Met, UGA=Trp); evolved separately, bacterial-like.
- What is a synonym codon? a) Stop codon b) Codon for same amino acid c) Anticodon d) Nonsense codon Answer: b) Codon for same amino acidReason: Due to degeneracy, e.g., CUU and CUC both Leu (synonyms).
- The genetic code is: a) Ambiguous b) Degenerate and unambiguous c) Overlapping d) Comma-less only Answer: b) Degenerate and unambiguousReason: Degenerate (multiple codons/AA); unambiguous (one codon=one AA); also comma-less, non-overlapping.
Section 3: Discovery of Genetic Code (MCQs 21-25)
- Who first deciphered UUU codon in 1961? a) Francis Crick b) Har Gobind Khorana c) Marshall Nirenberg d) Sydney Brenner Answer: c) Marshall NirenbergReason: Nirenberg & Matthaei used poly-U mRNA in cell-free system; produced poly-Phe, so UUU=Phe. Nobel 1968.
- Which experiment used synthetic copolymers to map all codons? a) Phage T4 frame-shift b) Khorana's repeating copolymers c) Nirenberg's cell-free d) Gamow's diamond code Answer: b) Khorana's repeating copolymersReason: Khorana synthesized poly-UUC etc., predicting codon sequences; confirmed full code. Shared Nobel 1968.
- Who proposed the wobble hypothesis? a) Nirenberg b) Khorana c) Francis Crick d) Watson Answer: c) Francis CrickReason: In 1966, Crick explained third-base wobble (e.g., I pairs U/C/A) for fewer tRNAs.
- What confirmed the triplet nature of the code? a) Poly-U experiment b) Crick & Brenner's frame-shift mutants c) Khorana synthesis d) All of above Answer: d) All of aboveReason: Frame-shifts in phages (1957) showed triplets; poly-U (1961) and synthesis (1960s) built on it.
- When was the full genetic code deciphered? a) 1953 b) 1961 c) 1967 d) 1970 Answer: c) 1967Reason: By 1967, experiments verified universality in diverse organisms, completing the map.
Section 4: tRNA Structure (MCQs 26-30)
- What is the secondary structure of tRNA? a) Double helix b) Cloverleaf c) L-shape d) Hairpin Answer: b) CloverleafReason: Base-pairing forms acceptor stem, anticodon loop, D-loop, TψC loop, variable loop—like a four-leaf clover.
- Where is the anticodon located in tRNA? a) 3' end b) Acceptor stem c) Anticodon loop d) D-loop Answer: c) Anticodon loopReason: 7-nt loop on 5' side; anticodon triplet base-pairs with mRNA codon for specificity.
- What sequence is at the 3' end of tRNA for amino acid attachment? a) AUG b) CCA c) UAC d) GGG Answer: b) CCAReason: Universal CCA accepts amino acid via ester bond; conserved across tRNAs.
- tRNA's tertiary structure resembles: a) Straight line b) L-shape c) Circle d) Helix Answer: b) L-shapeReason: D-arm + anticodon arm = one leg; acceptor + T-arm = other; ~7 nm, stabilized by Mg²⁺.
- What percentage of tRNA bases are modified? a) 1-5% b) 10-20% c) 50% d) 100% Reason: b) 10-20%Reason: Modifications like pseudouridine (ψ), inosine enhance stability, wobble pairing, and prevent degradation.
Section 5: rRNA Structure (MCQs 31-35)
- How many rRNA types in prokaryotic 70S ribosome? a) 2 b) 3 c) 4 d) 5 Answer: c) 4Reason: 16S (small), 23S + 5S (large); 23S has PTC.
- What is the eukaryotic small subunit rRNA? a) 16S b) 18S c) 23S d) 28S Answer: b) 18SReason: 18S in 40S subunit (~1,900 nt), homologous to prokaryotic 16S for decoding.
- Where is the peptidyl transferase center located? a) Small subunit b) Domain V of 23S/28S rRNA c) tRNA loop d) mRNA Answer: b) Domain V of 23S/28S rRNAReason: RNA-based catalysis in large subunit; no proteins involved—ribozyme proof.
- Eukaryotic large subunit has how many rRNAs? a) 1 b) 2 c) 3 d) 4 Answer: c) 3Reason: 28S, 5.8S, 5S in 60S; processed from 45S pre-rRNA in nucleolus.
- What Nobel Prize recognized ribosome structure? a) 1968 (code) b) 2009 (cryo-EM) c) 1953 (DNA) d) 1989 (PCR) Answer: b) 2009 (cryo-EM)Reason: Ramakrishnan, Steitz, Yonath for atomic rRNA details, confirming RNA world.
Section 6: Initiation, Elongation, Termination (MCQs 36-50)
- What binds first in prokaryotic initiation? a) Large subunit b) mRNA to 30S c) tRNA to A-site d) Release factors Answer: b) mRNA to 30SReason: 30S + mRNA (Shine-Dalgarno) + fMet-tRNA + IFs; then 50S joins.
- Eukaryotic initiation uses: a) Shine-Dalgarno b) 5' cap scanning c) Formyl group d) Rho factor Answer: b) 5' cap scanningReason: 40S + eIFs scans from 5' cap to Kozak-AUG; more regulated than prokaryotes.
- In elongation, what catalyzes peptide bond? a) EF-Tu b) Peptidyl transferase (rRNA) c) RF1 d) eIF2 Answer: b) Peptidyl transferase (rRNA)Reason: Ribozyme activity transfers chain from P-tRNA to A-tRNA.
- Translocation in elongation requires: a) EF-Tu-GTP b) EF-G-GTP c) IF2 d) RF3 Answer: b) EF-G-GTPReason: EF-G hydrolyzes GTP to ratchet ribosome, shifting tRNAs and mRNA by 3 nt.
- How many GTP per amino acid in elongation? a) 1 b) 2 c) 3 d) 0 Answer: b) 2Reason: 1 for EF-Tu (aa-tRNA binding), 1 for EF-G (translocation).
- Termination occurs when: a) AUG enters A-site b) Stop codon in A-site c) tRNA exits E-site d) Initiation factors bind Answer: b) Stop codon in A-siteReason: No tRNA for stops; RFs bind, hydrolyze chain-tRNA bond.
- Prokaryotic release factor for all stops? a) RF1 b) RF2 c) RF3 d) None; specific Answer: d) None; specificReason: RF1 (UAA/UAG), RF2 (UAA/UGA); RF3 aids GTP-dependent release.
- Eukaryotic termination uses: a) RF1 only b) eRF1 (all stops) + eRF3 c) IF3 d) EF-Tu Answer: b) eRF1 (all stops) + eRF3Reason: eRF1 recognizes all three stops; more unified than prokaryotes.
- Post-termination, what helps ribosome dissociation? a) EF-G b) IF3 (prok) or eIFs (euk) c) tRNA d) mRNA cap Answer: b) IF3 (prok) or eIFs (euk)Reason: Recycling factors split subunits for reuse.
- Direction of polypeptide growth? a) C to N b) N to C c) Random d) 5' to 3' Answer: b) N to CReason: Amino terminus (N) starts at initiator; new AAs add to carboxyl (C) end.
- Antibiotic blocking A-site entry? a) Streptomycin b) Tetracycline c) Puromycin d) Rifampicin Answer: b) TetracyclineReason: Binds 30S, prevents aa-tRNA to A-site; inhibits elongation.
- Initiation energy cost? a) 0 GTP b) 1 GTP c) 2 GTP d) ATP only Answer: b) 1 GTPReason: IF2/eIF2-GTP for subunit joining; hydrolyzed upon assembly.
- Termination energy? a) ATP b) 1 GTP (RF3/eRF3) c) 2 GTP d) None Answer: b) 1 GTP (RF3/eRF3)Reason: Aids RF binding and hydrolysis for release.
- Prok vs Euk difference in translation? a) Coupled with transcription in prok b) Larger ribosomes in prok c) No start codon in euk d) Faster in euk Answer: a) Coupled with transcription in prokReason: No nucleus in prokaryotes; translation starts on nascent mRNA.
- Error rate in translation? a) 1 in 10 b) 1 in 10,000 c) 1 in 1,000,000 d) 100% accurate Answer: b) 1 in 10,000Reason: Proofreading by synthetases, GTP hydrolysis, and wobble ensure high fidelity; mutations from errors cause diseases.
🧬 Interactive Protein Synthesis & Genetic Code Quiz (BS Level - 50 Questions)
Test your knowledge! Select an answer, click Submit for feedback, then Next. Progress tracked. Full 50 MCQs included.




+by+Blacklotus+Landscaping.jpg)


.jpg)


0 Comments